 Methods & Techniques
Methods & Techniques
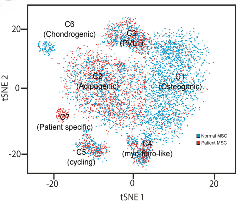
Experimental systems include CRISPR-edited cell lines and mouse models, cancer stem cells, cancer patient-derived organoids, and xenografts. Experimental approaches include genome analysis (single-cell sequencing, RNA-seq, ATAC-seq, CHiRP, CHiRP Mass Spec, PRO-seq, Cut and Run, Radical Seq, BioID, and chromosome conformation capture and Micro C), spatial transcriptomics (NanoString), multispectral imaging (CODEX), degron strategies, and analysis of patient-derived tissue specimens.